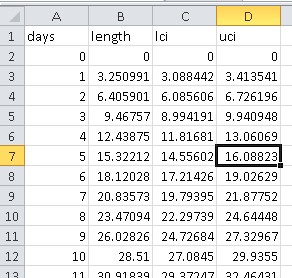
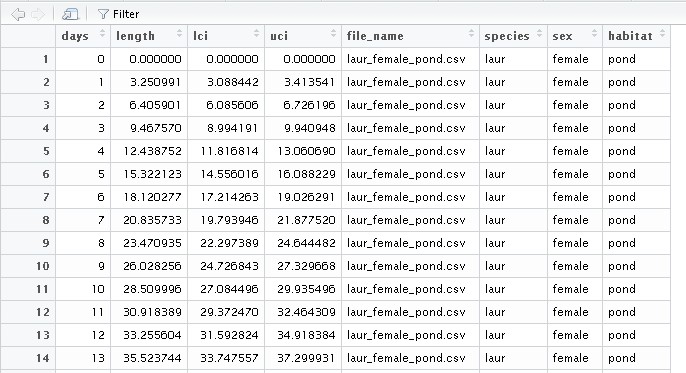
Mapping Australian electorates I've visited
/R packages are getting so specific that any idea can be relatively easily achieved in a short time. This time, I wanted to map the Australian electorates that I have visited. For no particular reason.
A quick Google found the eechidna package, which allows quick and easy mapping of Australian electorates in ggplot. data(nat_map) provides the polygons.
I then exported the 150 electorates so that I could create a variable for the electorates I've visited (this is time-consuming), and re-imported it. This is the point at which you could do some more interesting visualisations.
elec_lookup <- data.frame(ELECT_DIV = unique(nat_map$ELECT_DIV))
write.csv(elec_lookup, "elec_lookup.csv")
##Add visited electorates variable in your spreadsheet program of choice##
elec_lookup <- read.csv("elec_lookup.csv")
nat_map_2 <- merge(nat_map, elec_lookup, by="ELECT_DIV")
Time to create the map
cbPalette <- c("#999999", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7") ##Colourblind pallette
plot_aus <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "gray90", fill = NA, size = 2))
The obvious issue here is that the smaller city electorates are practically invisible. It's essentially impossible to tell that I've visited Melbourne!
I created some zoomed-in maps for the five largest cities. The hardest issue here is limiting the axes properly to not screw up the polygons:
plot_per <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
coord_cartesian(xlim = c(115, 117), ylim = c(-31.27, -32.5)) + # limit the axes using coord_cartesian
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect())Finally, I needed to arrange in a grid. This is relatively simple, but takes some trial-and-error to get the ratios right. I also created some rectangles to link the sub-maps by box colour.
Below is the full code:
librrary(ggplot2)
library(eechidna)
library(gridExtra)
data(nat_map)
elec_lookup <- data.frame(electorates = unique(nat_map$ELECT_DIV))
write.csv(elec_lookup, "elec_lookup.csv")
elec_lookup <- read.csv("elec_lookup.csv")
nat_map_2 <- merge(nat_map, elec_lookup, by="ELECT_DIV")
cbPalette <- c("#999999", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7")
plot_aus <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
geom_rect(aes(xmin = 115, xmax = 117, ymin = -31.27, ymax = -32.5), colour = "#009E73", fill = NA, size = 1.1) + ##Perth
geom_rect(aes(xmin = 138, xmax = 140, ymin = -34.27, ymax = -35.5), colour = "#F0E442", fill = NA, size = 1.1) + ##Adelaide
geom_rect(aes(xmin = 152, xmax = 154, ymin = -26.77, ymax = -28), colour = "#0072B2", fill = NA, size = 1.1) + ##Brisbane
geom_rect(aes(xmin = 150, xmax = 152, ymin = -32.54, ymax = -35), colour = "#D55E00", fill = NA, size = 1.1) + ##Sydney
geom_rect(aes(xmin = 144, xmax = 146, ymin = -37.27, ymax = -38.5), colour = "#CC79A7", fill = NA, size = 1.1) + ##Melbourne
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "gray90", fill = NA, size = 2))
##Perth##
plot_per <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
coord_cartesian(xlim = c(115, 117), ylim = c(-31.27, -32.5)) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "#009E73", fill = NA, size = 2))
##Adelaide##
plot_ade <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
coord_cartesian(xlim = c(138, 140), ylim = c(-34.27, -35.5)) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "#F0E442", fill = NA, size = 2))
##Brisbane##
plot_bri <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
coord_cartesian(xlim = c(152, 154), ylim = c(-26.5, -28)) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "#0072B2", fill = NA, size = 2))
##Sydney##
plot_syd <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
coord_cartesian(xlim = c(150, 152), ylim = c(-32.54, -35)) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "#D55E00", fill = NA, size = 2))
##Melbourne##
plot_mel <- ggplot(data=nat_map_2) +
geom_polygon(aes(x=long, y=lat, group=group, fill = as.factor(been)), colour="white") +
guides(fill = FALSE) +
scale_fill_manual(values=cbPalette) +
coord_cartesian(xlim = c(144, 146),ylim = c(-37.27, -38.5)) +
xlab("") +
ylab("") +
theme_classic() +
theme(axis.line=element_blank(),axis.text.x=element_blank(),
axis.text.y=element_blank(),axis.ticks=element_blank(),
panel.border = element_rect(colour = "#CC79A7", fill = NA, size = 2))
grid.arrange(plot_aus, plot_bri, plot_syd, plot_mel, plot_per, plot_ade, widths = c(2, 2, 2, 2, 2, 2),
layout_matrix = rbind(c(1, 1, 1, 1, 2, 2),
c(1, 1, 1, 1, 3, 3),
c(1, 1, 1, 1, 3, 3),
c(5, 5, 6, 6, 4, 4)))